grupa badawcza
Grupa badawcza wirusologii ilościowej
Tematyka badawcza
One of the main objectives of our laboratory is to study the establishment of HIV latency coupled with stochastic HIV transcription and the host functional genome. Using quantitative gnomic and machine-learning-based approaches tailored from the laboratory, we aim to deepen our grasp on how variations, with resolution of single viruses, in HIV genome integrity and transcription influence the configuration of HIV reservoirs. Two research lines aligning with this objective are conducted.
1. Role of HIV antisense transcripts (AST) in the establishment of latency
Although the existence of HIV AST and encoded proteins was first postulated by computer analyses in 1988, at present the fundamental mechanisms behind HIV AST are not fully understood. HIV antisense RNAs have been demonstrated in vitro to be capable of promoting the initiation and maintenance of HIV latency; however, HIV AST in vivo is poorly detected and the observations are contradictory, leaving uncertainty to conclude the mechanism of how HIV AST contributes to its pathogenesis. We characterize the possible threshold of the ratio between HIV sense and antisense transcription, determining the transcriptional phenotypes of HIV based on barcoded HIV and cellular clones possessing unique transcriptional phenotypes of HIV.
2. Distinguishable topology of the task-evoked functional genome networks in HIV reservoirs
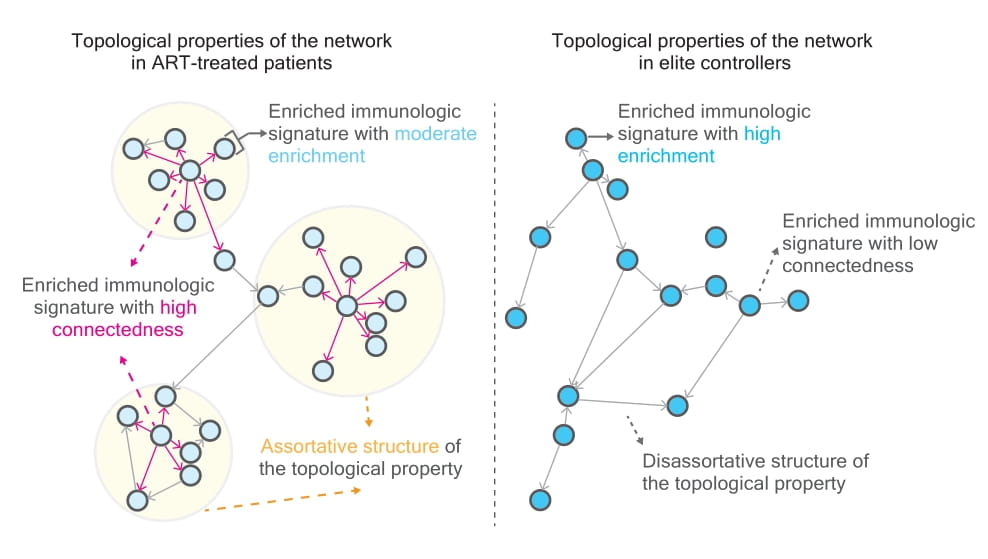
Our len to view HIV reservoir is that it can be represented as the topological (task-evoked) property of a network consisting of different communities of the genes targeted by HIV. Such communities are so-called immunologic signatures. HIV integration frequency within a network might be used as a proxy to define specific immune cell types and proinflammatory soluble factors, facilitating fine-tuning of the microenvironment of reservoirs.
To deepen our understanding of such heterogeneous HIV reservoirs and their functional implications, we pioneer the integration of a convergence approach to characterize the composition of HIV reservoirs. Based on graph-theoretical tools, we observe noticeable topological properties in networks, featuring immunologic signatures enriched by genes harboring intact and defective proviruses, when comparing antiretroviral therapy (ART)-treated HIV-1-infected individuals and elite controllers.
The key variable, the rich factor, plays a pivotal role in classifying distinct topological properties in networks. The host gene expression strengthens the accuracy of classification between elite controllers and ART-treated patients. Markov chain Monte Carlo modeling for the simulation of different graph networks demonstrated the presence of an intrinsic barrier between elite controllers and non-elite controllers. Notably, our work provides a prime example of leveraging genomic approaches alongside mathematical tools to unravel the complexities of HIV reservoirs.
Publications related to this research line:
Chen, H.-C. 2023. Vaccines DOI:10.3390/vaccines11020402
Więcek, K., and Chen, H.-C. 2023. iScience DOI:10.1016/j.isci.2023.108342
Wiśniewski, et al. 2024. iScience DOI:10.1016/j.isci.2024.11122
In our laboratory, we also study zoonotic viral infection. We study the linkage between the intrinsic tropisms of coronavirus variants and host domestication.
3. Identification of potential SARS-CoV-2 genetic markers resulting from host domestication
We develop a k–mer–based pipeline, namely the Pathogen Origin Recognition Tool using Enriched K–mers (PORT-EK) to identify genomic regions enriched in the respective hosts after the comparison of metagenomes of isolates between two host species.
Using it we successfully identify thousands of k–mers enriched in US white-tailed deer and betacoronaviruses while comparing them with human isolates. In addition, we demonstrate different coverage landscapes of k–mers enriched in deer and bats and unravel 144 mutations in enriched k–mers yielded from the comparison of viral metagenomes between bat and human isolates.
Additionally, we observe that the third position within a genetic codon is prone to mutations, resulting in a high frequency of synonymous mutations of amino acids harboring the same physicochemical properties as unaltered amino acids. Importantly, we are able to classify and predict the likelihood of host species based on the enriched k-mer counts.

Publications related to this research line:
Wiśniewski and Chen. 2024. BioRxiv DOI:10.1101/2024.07.27.605454
